Introduction to DEA Fractional Cover 
Sign up to the DEA Sandbox to run this notebook interactively from a browser
Compatibility: Notebook currently compatible with both the
NCIandDEA SandboxenvironmentsProducts used: ga_ls_fc_3, ga_ls_wo_3
Background
Fractional cover data can be used to identify large scale patterns and trends and inform evidence based decision making and policy on topics including wind and water erosion risk, soil carbon dynamics, land management practices and rangeland condition.
This information is used by policy agencies, natural and agricultural land resource managers, and scientists to monitor land conditions over large areas over long time frames.
What this product offers
Fractional Cover (FC), developed by the Joint Remote Sensing Research Program, is a measurement that splits the landscape into three parts, or fractions:
green (leaves, grass, and growing crops)
brown (branches, dry grass or hay, and dead leaf litter)
bare ground (soil or rock)
DEA uses Fractional Cover to characterise every 30 m square of Australia for any point in time from 1987 to today.
Applications
Fractional cover provides valuable information for a range of environmental and agricultural applications, including:
soil erosion monitoring
land surface process modelling
land management practices (e.g. crop rotation, stubble management, rangeland management)
vegetation studies
fuel load estimation
ecosystem modelling
land cover mapping
Description
This notebook will demonstrate how to load DEA Fractional Cover using Digital Earth Australia. Topics covered include:
Inspecting the products and measurements available in the datacube
Loading DEA Fractional Cover for an example location
Plotting fractional cover as false colour images
Inspecting unmixing error outputs
Masking out missing or invalid data and unclear or wet pixels, and using this to track percentages of green and brown vegetation and bare soil over time
Note: Visit the DEA Fractional Cover product documentation for detailed technical information including methods, quality, and data access.
Getting started
To run this analysis, run all the cells in the notebook, starting with the “Load packages” cell.
Load packages
Import Python packages that are used for the analysis.
[1]:
import datacube
from datacube.utils import masking
import matplotlib.pyplot as plt
from dea_tools.datahandling import wofs_fuser
from dea_tools.plotting import rgb, plot_wo
Connect to the datacube
Connect to the datacube so we can access DEA data.
[2]:
dc = datacube.Datacube(app='DEA_Fractional_Cover')
Available products and measurements
List products available in Digital Earth Australia
We can use datacube’s list_products functionality to inspect DEA Fractional Cover products that are available in Digital Earth Australia. The table below shows the product name that we will use to load data, and a brief description of the product.
[3]:
# List DEA Fractional Cover products available in DEA
dc_products = dc.list_products()
dc_products.loc[['ga_ls_fc_3']]
[3]:
| name | description | license | default_crs | default_resolution | |
|---|---|---|---|---|---|
| name | |||||
| ga_ls_fc_3 | ga_ls_fc_3 | Geoscience Australia Landsat Fractional Cover ... | CC-BY-4.0 | EPSG:3577 | Resolution(x=30, y=-30) |
List measurements
We can inspect the contents of the DEA Fractional Cover product using datacube’s list_measurements functionality. The table below lists each of the measurements available in the product, which represent unique data variables that provide information about the vegetation and bare soil cover in each pixel:
pv: The fractional cover of green vegetationnpv: The fractional cover of non-green vegetationbs: The fractional cover of bare soilue: The fractional cover unmixing error
The table also provides information about the measurement data types, units, nodata value and other technical information about each measurement.
[4]:
dc_measurements = dc.list_measurements()
dc_measurements.loc[['ga_ls_fc_3']]
[4]:
| name | dtype | units | nodata | aliases | flags_definition | ||
|---|---|---|---|---|---|---|---|
| product | measurement | ||||||
| ga_ls_fc_3 | bs | bs | uint8 | percent | 255 | [bare] | NaN |
| pv | pv | uint8 | percent | 255 | [green_veg] | NaN | |
| npv | npv | uint8 | percent | 255 | [dead_veg] | NaN | |
| ue | ue | uint8 | 1 | 255 | [err] | NaN |
Loading data
Now that we know what products and measurements are available for the products, we can load data from Digital Earth Australia for an example location:
[5]:
# Set up a region to load data
time = ('1993-10-15', '1993-11-20')
query = {
'y': (-34.06813, -34.22099),
'x': (139.83315, 140.05151),
'time': time,
}
# Load DEA Fractional Cover data from the datacube
fc = dc.load(product='ga_ls_fc_3',
measurements=['bs', 'pv', 'npv', 'ue'],
output_crs='EPSG:32754',
resolution=(-30, 30),
group_by='solar_day',
**query)
We can now view the data that we loaded. The measurements listed under Data variables should match the measurements displayed in the previous List measurements step.
[6]:
fc
[6]:
<xarray.Dataset> Size: 6MB
Dimensions: (time: 4, y: 573, x: 678)
Coordinates:
* time (time) datetime64[ns] 32B 1993-10-21T23:55:41.593257 ... 199...
* y (y) float64 5kB 6.23e+06 6.23e+06 ... 6.213e+06 6.213e+06
* x (x) float64 5kB 3.923e+05 3.924e+05 ... 4.126e+05 4.126e+05
spatial_ref int32 4B 32754
Data variables:
bs (time, y, x) uint8 2MB 16 19 16 24 20 29 ... 17 20 16 23 26 13
pv (time, y, x) uint8 2MB 51 52 53 52 50 38 ... 32 28 30 26 20 31
npv (time, y, x) uint8 2MB 32 27 29 23 29 32 ... 50 51 53 50 53 55
ue (time, y, x) uint8 2MB 13 13 12 12 11 9 9 ... 12 12 13 11 11 13
Attributes:
crs: EPSG:32754
grid_mapping: spatial_refPlotting data
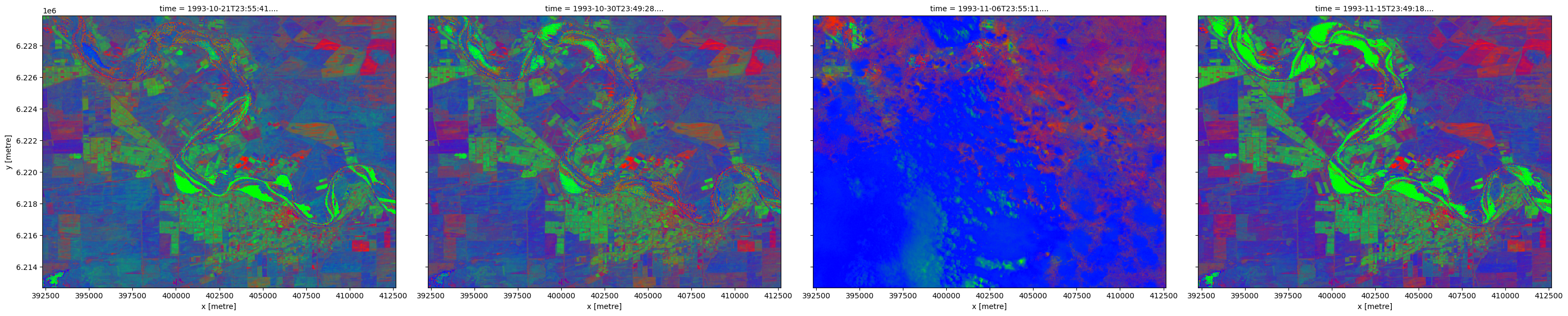
We can plot each FC variable in our dataset (i.e. ['bs', 'pv', 'npv']) using the rgb function. This will create a false colour view of the data where shades of green, blue and red represent varying proportions of vegetation and bare soil cover:
Green: green vegetation (
pv)Blue: brown (i.e. ‘non-green’) vegetation (
npv)Red: bare soil (
bs)
The resulting images show agricultural fields containing high proportions of green vegetation cover, surrounded by areas dominated by brown vegetation and bare soil. Note that the third image contains dense clouds which are erroneously mapped as non-growing vegetation.
Note: Fractional cover values range between 0 and 100%, but due to model uncertainties and the limitations of the training data, some areas may show cover values in excess of 100%. These areas can either be excluded or treated as equivalent to 100%.
[7]:
# Plot DEA Fractional Cover data as a false colour RGB image
rgb(fc, bands=['bs', 'pv', 'npv'], col='time')
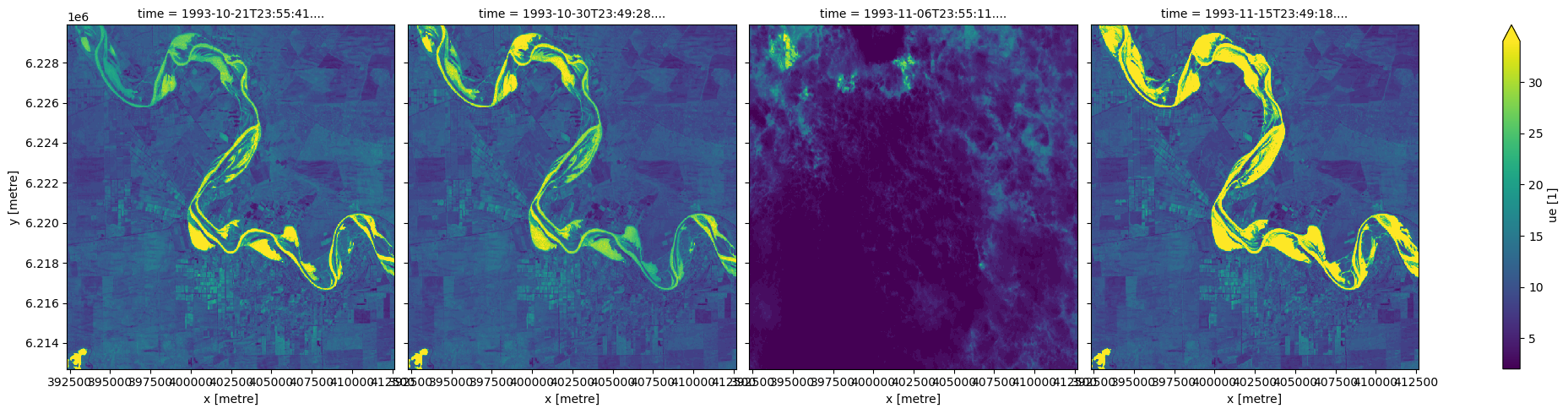
We can also visualise the ‘unmixing error’ (ue) for each of our DEA Fractional Cover observations. High unmixing error values (bright colours below) represent areas of higher model uncertainty (e.g. areas of water, cloud, cloud shadow or soil types/colours that were not included in the model training data). This data can be useful for removing uncertain pixels from an analysis.
In this example, wet pixels associated with a river have relatively high unmixing errors.
[8]:
# Plot unmixing error using `robust=True` to drop outliers and improve contrast
fc.ue.plot(col='time', robust=True, size=5)
[8]:
<xarray.plot.facetgrid.FacetGrid at 0x7f56b55a9720>
Note: For more technical information about the accuracy and limitations of DEA Fractional Cover, refer to the Details tab of the official Geoscience Australia DEA Fractional Cover product description.
Example application: tracking changes in vegetation cover and bare soil over time
The following section will demonstrate a simple analysis workflow based on DEA Fractional Cover. In this example, we will process our loaded FC data so that we can consistently track the changing proportions of green vegetation, brown vegetation and bare soil over time.
Setting nodata
As the first step in our analysis, we need to set nodata pixels to NaN. This ensures that missing data is dealt with correctly in any future calculations.
[9]:
# Replace all nodata values with `NaN`
fc = masking.mask_invalid_data(fc)
Applying a cloud and water mask
In the images we plotted earlier, you may have noticed the third panel is affected by cloud. This can cause the fractional cover algorithm to produce misleading results. FC will also produce poor results over water, causing erroneous values for green vegetation to appear within wet pixels (e.g. the fourth panel above).
To track FC reliably over time, we need to remove these potentially inaccurate pixels. One of the easiest ways to do this is to load data from the DEA Water Observations product that identifies wet and unclear pixels (e.g. cloud or cloud shadow) in the landscape.
In the next cell, we load DEA Water Observations data into the extents of our DEA Fractional Cover data using dc.load()’s like argument.
Note: For more details about loading data, refer to the Introduction to loading data notebook.
[10]:
# Load DEA Water Observations data from the datacube
wo = dc.load(product='ga_ls_wo_3',
group_by='solar_day',
fuse_func=wofs_fuser,
like=fc)
# Plot the loaded water observations
plot_wo(wo.water, col='time', size=5)
[10]:
<xarray.plot.facetgrid.FacetGrid at 0x7f56b6ebe7d0>
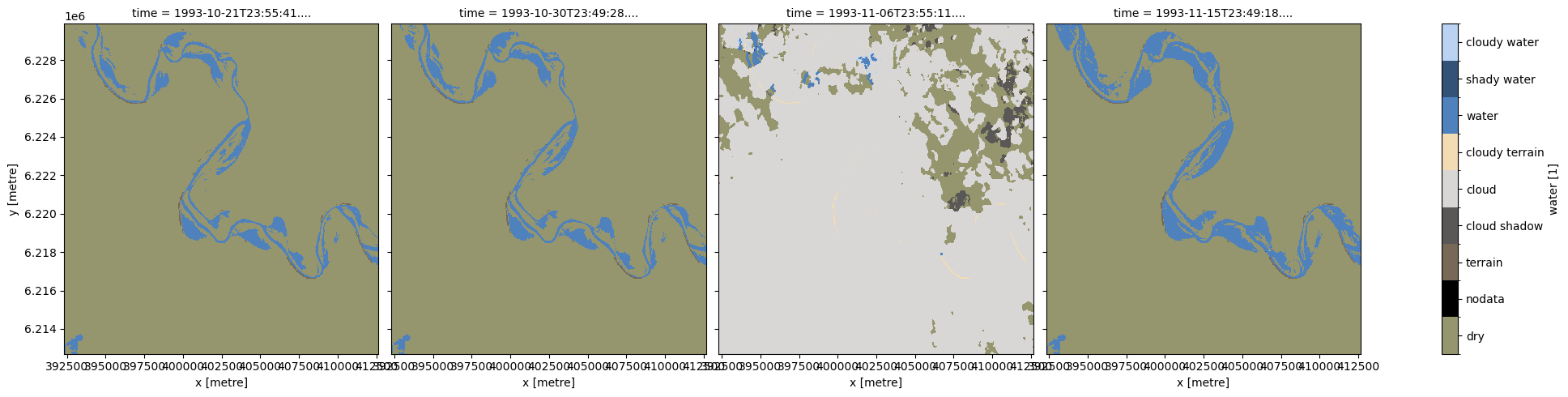
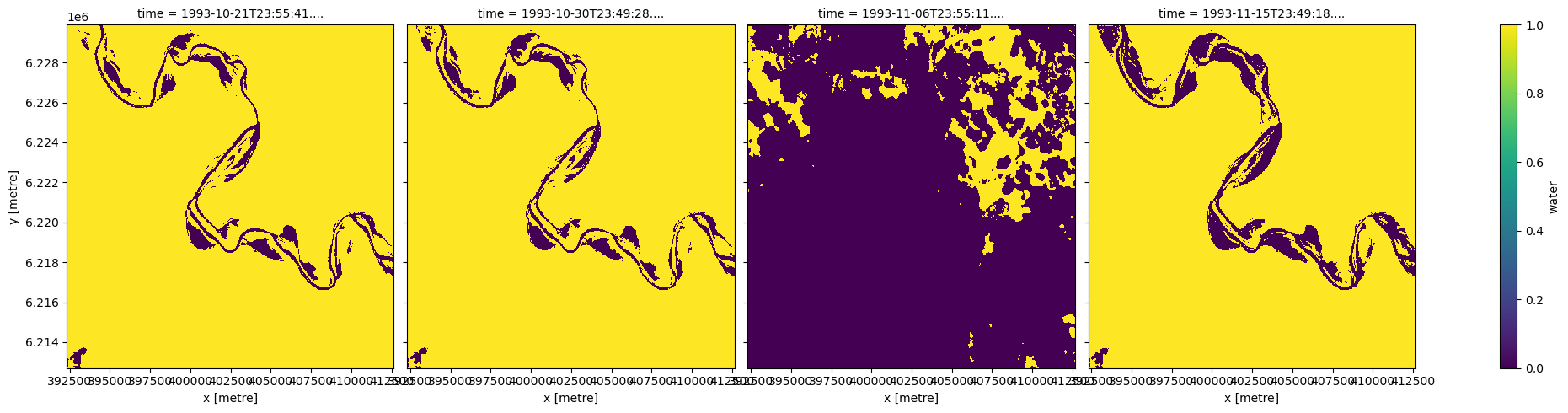
This plot shows that all images contain water, and the third image contains large amounts of cloud and cloud shadow. To remove these pixels, we first create a binary mask where True (yellow) represents clear and dry pixels, and False (purple) represents wet, cloudy or shadowy pixels.
Note: For a detailed guide to using DEA Water Observations data to mask data, see the DEA Water Observations notebook.
[11]:
# Keeping only dry, non-cloudy pixels
wo_mask = masking.make_mask(wo.water, dry=True)
wo_mask.plot(col='time', size=5)
[11]:
<xarray.plot.facetgrid.FacetGrid at 0x7f56b48cc130>
Using this mask, we can now remove any wet and unclear pixels from our original DEA Fractional Cover data. Note that these pixels now appear white as they have been replaced with NaN.
[12]:
# Set any unclear or wet pixel to `NaN`
fc_masked = fc.where(wo_mask)
# Plot the masked fractional cover data
rgb(fc_masked, bands=['bs', 'pv', 'npv'], col='time')
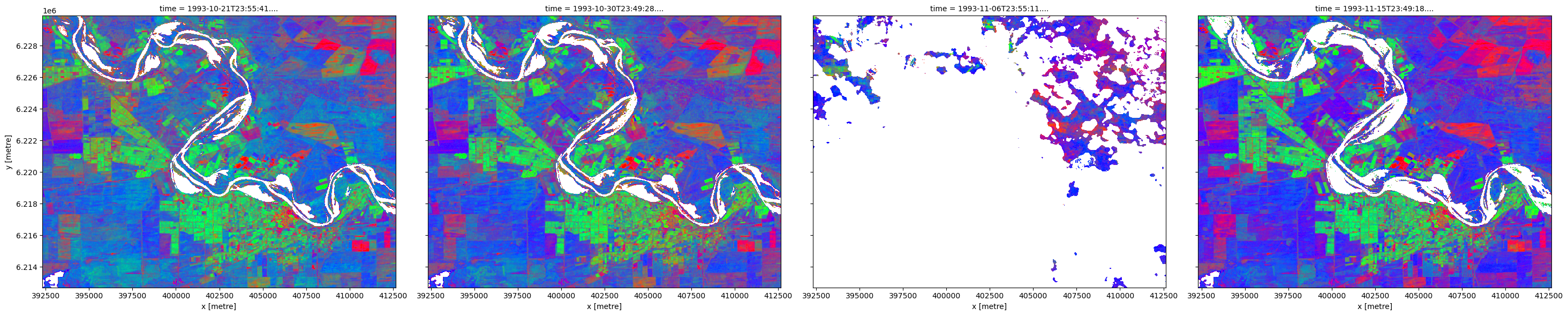
Dropping poorly observed scenes
In the image above, we can see that the third observation was mostly obscured by cloud, leaving very little usable data. So that this doesn’t lead to unrepresentative statistics, we can keep only observations that had (for example) less than 50% nodata pixels.
[13]:
# Calculate the percent of nodata pixels in each observation
percent_nodata = fc_masked.pv.isnull().mean(dim=['x', 'y'])
# Use this to filter to observations with less than 50% nodata
fc_masked = fc_masked.sel(time=percent_nodata < 0.5)
# The data now contains only three observations
fc_masked
[13]:
<xarray.Dataset> Size: 19MB
Dimensions: (time: 3, y: 573, x: 678)
Coordinates:
* time (time) datetime64[ns] 24B 1993-10-21T23:55:41.593257 ... 199...
* y (y) float64 5kB 6.23e+06 6.23e+06 ... 6.213e+06 6.213e+06
* x (x) float64 5kB 3.923e+05 3.924e+05 ... 4.126e+05 4.126e+05
spatial_ref int32 4B 32754
Data variables:
bs (time, y, x) float32 5MB 16.0 19.0 16.0 24.0 ... 23.0 26.0 13.0
pv (time, y, x) float32 5MB 51.0 52.0 53.0 52.0 ... 26.0 20.0 31.0
npv (time, y, x) float32 5MB 32.0 27.0 29.0 23.0 ... 50.0 53.0 55.0
ue (time, y, x) float32 5MB 13.0 13.0 12.0 12.0 ... 11.0 11.0 13.0
Attributes:
crs: EPSG:32754
grid_mapping: spatial_refCalculating average fractional cover over time
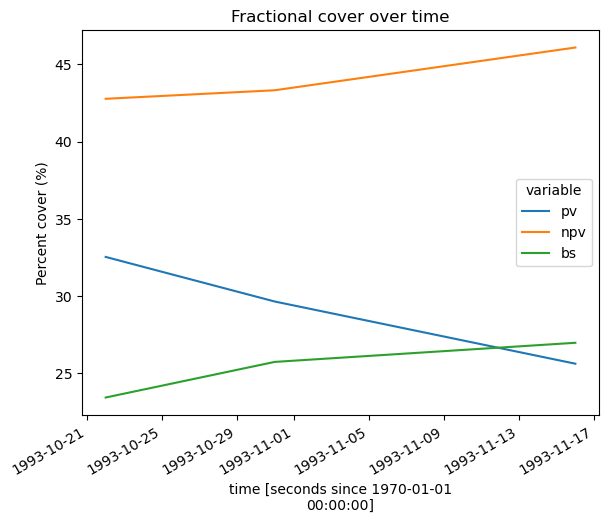
Now that our FC data has had nodata values and cloud, shadow and water pixels set to NaN and we have dropped unrepresentative observations, we can reliably track how average proportions of green and brown vegetation and bare soil have changed over time across our entire study area. We can then plot this as a line chart, showing that green vegetation (pv) has consistently decreased over time at this location, and brown vegetation (npv) and bare soil (bs) have increased.
[14]:
# Calculate average fractional cover for `bs`, `pv` and `npv` over time
fc_through_time = fc_masked[['pv', 'npv', 'bs']].mean(dim=['x', 'y'])
# Plot the changing proportions as a line graph
fc_through_time.to_array().plot.line(hue='variable', size=5)
plt.title('Fractional cover over time')
plt.ylabel('Percent cover (%)');
Additional information
License: The code in this notebook is licensed under the Apache License, Version 2.0. Digital Earth Australia data is licensed under the Creative Commons by Attribution 4.0 license.
Contact: If you need assistance, please post a question on the Open Data Cube Discord chat or on the GIS Stack Exchange using the open-data-cube tag (you can view previously asked questions here). If you would like to report an issue with this notebook, you can file one on
GitHub.
Last modified: November 2025
Compatible datacube version:
[15]:
print(datacube.__version__)
1.9.10
Tags
Tags: NCI compatible, sandbox compatible, DEA products, ga_ls_fc_3, ga_ls_wo_3, rgb, plot_wo, fractional cover